|
Cancer - Prostate(구연)
|
(E-098)
|
|
|
한국 남성의 전립선암 예측을 위한 다중유전자 위험 점수의 유용성 평가 |
분당서울대학교병원 비뇨의학과
서울대학교 의과대학 비뇨의학교실
프로카젠¹ |
| 오종진, 송상헌, 예창희, 김정권, 이학민, 이상철, 홍성규, 변석수, 김은애¹, 우은진¹ |
BACKGROUND: To evaluate an aggregate influence of prostate cancer (PCa) susceptibility variants on the development of PCa in Korean men by using polygenic risk score (PRS) approach.
METHODS: An analysis of 1,001 cases of PCa and 2,641 controls was performed to: i) identify potential PCa-related risk loci in Koreans and ii) validate the cumulative association between these loci and PCa using PRS. Subgroup analyses based on risk stratification were conducted to better characterize the potential correlation to key PCa-related clinical outcomes (e.g., Gleason score, prostate specific antigen levels). The results were replicated using 514 cases of PCa and 548 controls from an independent cohort.
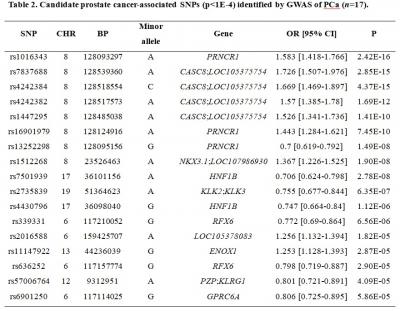
RESULTS: Genome-wide association analysis from our discovery cohort revealed 17 candidate SNPs associated with PCa showing statistical significance of p<1.0×10-4. Seven variants were located at 8q24.21 (rs1016343, rs16901979 and rs13252298 in PRNCR1; rs7837688, rs4242384, rs4242382 and rs1447295 in CASC8). Two variants located within HNF1B (rs7501939 and rs4430796) had a significant negative association with PCa risk (OR=0.706 and 0.747, p=2.78×10-8 and 1.12×10-6, respectively). Of the 9 independent SNPs that remained after LD pruning, the top 8 SNPs best predicted PCa risk with an AUC of 0.613 (95% CI: 0.58-0.647). Those with top 20% risk of PRS had an approximately two-fold increased risk of developing PCa compared with those having average polygenic risk.
CONCLUSIONS: Seventeen PCa-risk variants in Korean men were identified; PRSs of a subset of these variants could help predict PCa. The addition of individually calculated PRSs improved the accuracy of predicting PCa.
|
 |
|
keywords : prostate cancer, polygenic risk score, prediction |
|

